Oliveira, S. M. D., & Densmore, D. (2022, September 2). Hardware, software, and Wetware Codesign environment for Synthetic Biology. BioDesign Research. Retrieved from https://spj.sciencemag.org/journals/bdr/2022/9794510/
Landaverde L;Turcinovic J;Doucette-Stamm L;Gonzales K;Platt J;Connor JH;Klapperich C; (n.d.). Comparison of binaxnow and SARS-COV-2 qrt-PCR detection of the omicron variant from matched Anterior Nares swabs. Microbiology spectrum. Retrieved from https://pubmed.ncbi.nlm.nih.gov/36255297/
Turcinovic J;Kuhfeldt K;Sullivan M;Landaverde L;Platt JT;Doucette-Stamm L;Hanage WP;Hamer DH;Klapperich C;Landsberg HE;Connor JH; (n.d.). Linking contact tracing with genomic surveillance to deconvolute SARS-COV-2 transmission on a university campus. iScience. Retrieved from https://pubmed.ncbi.nlm.nih.gov/36246573/

Figure 1 from Oliveira, S. M. D., & Densmore, D. (2022). Hardware, software, and wetware codesign environment for synthetic biology. BioDesign Research. Retrieved from https://spj.sciencemag.org/journals/bdr/2022/9794510/

Bouton TC;Atarere J;Turcinovic J;Seitz S;Sher-Jan C;Gilbert M;White L;Zhou Z;Hossain MM;Overbeck V;Doucette-Stamm L;Platt J;Landsberg HE;Hamer DH;Klapperich C;Jacobson KR;Connor JH; (n.d.). Viral Dynamics of Omicron and delta SARS-COV-2 variants with implications for timing of release from isolation: A longitudinal cohort study. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. Retrieved from https://pubmed.ncbi.nlm.nih.gov/35737948/
Kuhfeldt K;Turcinovic J;Sullivan M;Landaverde L;Doucette-Stamm L;Hamer DH;Platt JT;Klapperich C;Landsberg HE;Connor JH; (n.d.). Examination of SARS-COV-2 in-class transmission at a large urban university with public health mandates using epidemiological and genomic methodology. Retrieved from https://pubmed.ncbi.nlm.nih.gov/35930286/
Petros BA;Turcinovic J;Welch NL;White LF;Kolaczyk ED;Bauer MR;Cleary M;Dobbins ST;Doucette-Stamm L;Gore M;Nair P;Nguyen TG;Rose S;Taylor BP;Tsang D;Wendlandt E;Hope M;Platt JT;Jacobson KR;Bouton T;Yune S;Auclair JR;Landaverde L;Klapperich CM;Hamer DH;Hana. (n.d.). Early introduction and rise of the omicron sars-COV-2 variant in highly vaccinated university populations. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. Retrieved from https://pubmed.ncbi.nlm.nih.gov/35616119/
Figure 1 from Kuhfeldt, K., Turcinovic, J., Sullivan, M., Landaverde, L., Doucette-Stamm, L., Hamer, D. H., Platt, J. T., Klapperich, C., Landsberg, H. E., & Connor, J. H. (n.d.). Examination of SARS-CoV-2 in-class transmission at a large urban university with public health mandates using epidemiological and genomic methodology. Retrieved from https://pubmed.ncbi.nlm.nih.gov/35930286/
Bouton TC;Atarere J;Turcinovic J;Seitz S;Sher-Jan C;Gilbert M;White L;Zhou Z;Hossain MM;Overbeck V;Doucette-Stamm L;Platt J;Landsberg HE;Hamer DH;Klapperich C;Jacobson KR;Connor JH; (n.d.). Viral Dynamics of Omicron and delta SARS-COV-2 variants with implications for timing of release from isolation: A longitudinal cohort study. medRxiv : the preprint server for health sciences. from https://pubmed.ncbi.nlm.nih.gov/35411341/
Landaverde, L., McIntyre, D., Robson, J., Fu, D., Ortiz, L., Chen, R., Oliveira, S.M.D., [...] (2021) Detailed Overview of the Buildout and Integration of an Automated High-Throughput CLIA Laboratory for SARS-CoV-2 Testing on a Large Urban Campus. medRxiv. doi:10.1101/2021.09.13.21263214.
Hamer, D.H., [...], Densmore, D., Brown, R.A. (2021) Assessment of a COVID-19 Control Plan on an Urban University Campus During a Second Wave of the Pandemic. JAMA Netw Open. 2021;4. doi:10.1001/jamanetworkopen.2021.16425

Figure 1 from Bouton, T. C., Atarere, J., Turcinovic, J., Seitz, S., Sher-Jan, C., Gilbert, M., White, L., Zhou, Z., Hossain, M. M., Overbeck, V., Doucette-Stamm, L., Platt, J., Landsberg, H. E., Hamer, D. H., Klapperich, C., Jacobson, K. R., & Connor, J. H. (n.d.). Viral dynamics of Omicron and Delta SARS-CoV-2 variants with implications for timing of release from isolation: A longitudinal cohort study. medRxiv. Retrieved from https://pubmed.ncbi.nlm.nih.gov/35411341/
Chen, R., Emery, N.J., Pavan, M., Oliveira, S.M.D. (2020) Laboratory Protocol Automation: A Modular DNA Assembly and Bacterial Transformation Case Study. In Proceedings of 12th IWBDA (IWBDA2020), Worcester, MA, Aug. 3-5. https://www.iwbdaconf.org/2020/docs/IWBDA2020Proceedings.pdf
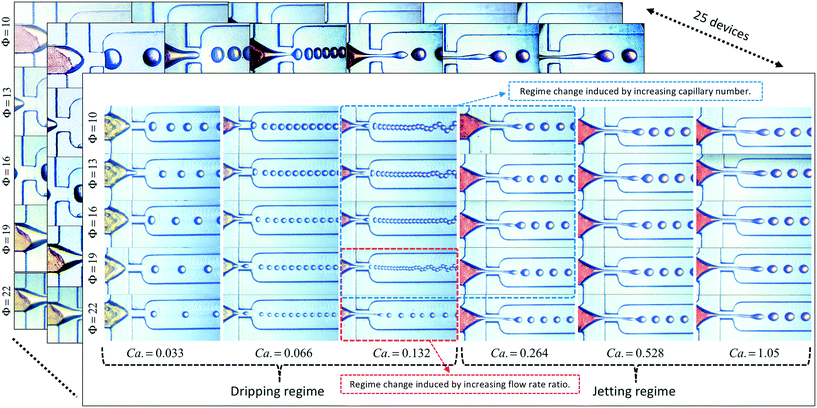
Lashkaripour, A., Rodriguez, C., Ortiz, L., and Densmore, D. (2019). Performance tuning of microfluidic flow-focusing droplet generators. Lab on a Chip, 19(6), 1041-1053. https://pubs.rsc.org/en/content/articlelanding/2019/lc/c8lc01253a/unauth#!divAbstract
Sanka, R., Lippai, J., Samarasekera, D., Nemsick, D., and Densmore, D. (2019). 3DμF - Interactive Design Environment for Continuous Flow Microfluidic Devices. Scientific Reports 9(1). https://doi.org/10.1038/s41598-019-45623-z
Figure 1 from Bouton, T. C., Atarere, J., Turcinovic, J., Seitz, S., Sher-Jan, C., Gilbert, M., White, L., Zhou, Z., Hossain, M. M., Overbeck, V., Doucette-Stamm, L., Platt, J., Landsberg, H. E., Hamer, D. H., Klapperich, C., Jacobson, K. R., & Connor, J. H. (n.d.). Viral dynamics of Omicron and Delta SARS-CoV-2 variants with implications for timing of release from isolation: A longitudinal cohort study. medRxiv. Retrieved from https://pubmed.ncbi.nlm.nih.gov/35411341/
Hillson, N., Caddick, M., [...], Freemont, P.S. (2019). Building a Global Alliance of Biofoundries. Nature Communications, 10:2040. https://www.nature.com/articles/s41467-019-10079-2
Walsh III, D.I., Pavan, M., Ortiz, L., Wick, S., Bobrow, J., Guido, N.J., Leinicke, S., Fu, D., Pandit, S., Qin, L., Carr, P.A., Densmore, D. (2019). Standardizing automated DNA assembly: best practices, metrics, and protocols using robots. SLAS TECHNOLOGY: Translating Life Sciences Innovation, 24(3), 282-290. https://journals.sagepub.com/doi/abs/10.1177/2472630318825335
Sanka, R., Crites, B., McDaniel, J., Brisk, P., and Densmore, D. (2019). Specification, Integration, and Benchmarking of Continuous Flow Microfluidic Devices: Invited Paper. In 2019 IEEE/ACM International Conference on Computer-Aided Design (ICCAD), 1–8. https://doi.org/10.1109/ICCAD45719.2019.8942171

Figure 3 from Walsh III, D.I., Pavan, M., Ortiz, L., Wick, S., Bobrow, J., Guido, N.J., Leinicke, S., Fu, D., Pandit, S., Qin, L., Carr, P.A., Densmore, D. (2019). Standardizing automated DNA assembly: best practices, metrics, and protocols using robots. SLAS TECHNOLOGY: Translating Life Sciences Innovation, 24(3), 282-290. https://journals.sagepub.com/doi/abs/10.1177/2472630318825335

Lashkaripour, A., Silva, R., and Densmore, D. (2018). Desktop micro-milled microfluidics. Microfluidics and Nanofluidics, 22:31. https://link.springer.com/article/10.1007/s10404-018-2048-2
Ortiz, L., Pavan, M., McCarthy, L., Timmons, J., Densmore, D. (2017). Automated Robotic Liquid Handling Assembly of Modular DNA Devices. J. Vis. Exp. (), e54703. https://www.jove.com/video/54703/automated-robotic-liquid-handling-assembly-of-modular-dna-devices
Silva, R., Dow, P., Dubay, R., Lissandrello, C., Holder, J., Densmore, D., & Fiering, J. (2017). Rapid prototyping and parametric optimization of plastic acousto-fluidic devices for blood–bacteria separation. Biomedical Microdevices, 19(3), 70. https://link.springer.com/article/10.1007/s10544-017-0210-3
Figure 1 from Lashkaripour, A., Silva, R., & Densmore, D. (2018). Desktop micro-milled microfluidics. Microfluidics and Nanofluidics, 22, 31. https://link.springer.com/article/10.1007/s10404-018-2048-2
Silva, R., Bhatia, S., and Densmore, D. (2016). A reconfigurable continuous-flow fluidic routing fabric using a modular, scalable primitive. Lab on a Chip 16.14: 2730-2741. https://pubs.rsc.org/en/content/articlehtml/2016/lc/c6lc00477f
Huang, H., and Densmore, D. (2014). Fluigi: Microfluidic device synthesis for synthetic biology. ACM Journal on Emerging Technologies in Computing Systems (JETC) 11.3: 26. https://dl.acm.org/doi/abs/10.1145/2660773
Huang, H., and Densmore, D. (2014). Integration of microfluidics into the synthetic biology design flow. Lab on a Chip 14.18 (2014): 3459-3474. https://pubs.rsc.org/en/content/articlehtml/2014/lc/c4lc00509k
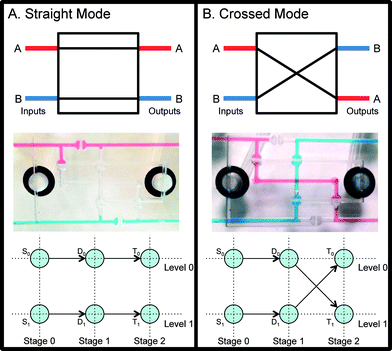
Figure 2 from Silva, R., Bhatia, S., & Densmore, D. (2016). A reconfigurable continuous-flow fluidic routing fabric using a modular, scalable primitive. Lab on a Chip, 16(14), 2730–2741. https://pubs.rsc.org/en/content/articlehtml/2016/lc/c6lc00477f